All published articles of this journal are available on ScienceDirect.
Antibiotic Susceptibility Patterns of Biofilm-producing Nosocomial Non-coagulase Staphylococcus Isolates from Clinical Sources in Iraq
Abstract
Introduction
Coagulase-negative staphylococci (CoNS), like S. epidermidis and S. haemolyticus, are skin commensals that can cause infections. Their biofilms increase resistance and persistence. This study assessed biofilm formation, antibiotic resistance, and genetic diversity of local CoNS using MLST to aid regional surveillance.
Methods
In this study, samples were collected, and the species were identified using the VITEK 2 Compact system. A Kirby-Bauer disc diffusion test was carried out to detect antimicrobial susceptibility, and the biofilm production was tested with a microtiter plate. Multilocus sequence typing (MLST) was performed, followed by data processing using the Oxford scheme to categorize the isolates.
Results
Among 150 clinical samples, 13.3% were S. epidermidis and 20% S. haemolyticus. Coagulase-negative staphylococci (CoNS) showed 100% resistance to amoxiclav, cefoxitin, methicillin, amoxicillin, and ceftriaxone. High resistance (66–96%) was seen for several antibiotics, with 76% classified as multidrug-resistant (MDR). All CoNS formed biofilms (58% strong). MLST revealed diverse sequence types (STs), including newly reported ST23 and ST59 for S. epidermidis, and ST24, ST35, and ST109 for S. haemolyticus in Iraq.
Discussion
Multidrug-resistant CoNS isolates showed high biofilm formation and mecA gene presence, indicating significant clinical infection and resistance challenges.
Conclusion
This study provides valuable data on the prevalence, antibiotic resistance, and genetic diversity of S. epidermidis and S. haemolyticus strains in Anbar, Iraq. It also reports several novel MLST sequence types for the first time in the region, including unique strain IDs for both species, contributing to improved local epidemiological knowledge and surveillance efforts.
1. INTRODUCTION
Coagulase-negative staphylococci (CoNS), including Staphylococcus epidermidis and Staphylococcus haemolyticus, are commonly present on the skin and mucous membranes of humans and animals [1].
They are acknowledged as opportunistic pathogens, with Staphylococcus epidermidis being the primary coagulase-negative Staphylococcus (CoNS) responsible for healthcare-associated infections (HAIs) [2]. HAIs, previously known as nosocomial or hospital-acquired infections, are infections acquired by patients during medical treatment in healthcare settings. They are a major contributor to illness and death among hospitalized patients [3]. According to the World Health Organization (WHO), approximately 7% of hospitalized patients in developed countries and 10% in developing countries are estimated to acquire at least one healthcare-associated infection (HAI) at any given time [4]. CoNS are primarily recognized as commensals on human and animal bodies, but they can also act as opportunistic pathogens. The composition of commensal skin flora varies by body site, reflecting the differing environmental conditions at each location [5]. It is noteworthy that biofilm-associated infections resist responding to different antimicrobial agents, the immune system, as well as harsh conditions; thus, biofilm formation is an important characteristic in the progression of pathogenicity. Biofilm-forming isolates contribute to infections associated with catheters and other medical equipment [6]. Biofilm-producing S. haemolyticus is a major cause of bacteremia, particularly in catheter-associated and nosocomial infections. Biofilm formation by S. haemolyticus is a complex process that is further enhanced in the presence of antimicrobial agents [7]. These biofilm-producing CoNS are increasingly reported to be resistant to “methicillin” and various antibiotic classes, including lincosamides and macrolides. This growing resistance poses a significant challenge in treating clinical infections, complicating antibiotic selection for patient care [8]. The most commonly isolated MR-CoNS species in hospitals are S. epidermidis and S. haemolyticus [9].
MLST is a molecular typing technique employed for bacteria identification and classification. It relies on analyzing DNA sequences from seven highly conserved housekeeping genes essential for cellular function [10]. These gene sequences are amplified via PCR and then compared against a reference database of known MLST types, allowing for the identification and assignment of the isolate to an MLST type. This classification offers insights into the bacterium's genetic lineage and potential pathogenicity [11]. This study aimed to identify prevalent local isolates and distinguish infectious pathogen strains using MLST to enhance epidemiological surveillance and strain source determination. Additionally, it investigated biofilm formation and antibiotic susceptibility patterns in Staphylococcus isolates. Given its role as a significant human pathogen, particularly in immunocompromised individuals, long-term hospitalized patients, and those with severe illnesses, understanding its genetic diversity and resistance profiles is crucial for effective infection control.
2. MATERIALS AND METHODS
2.1. Bacterial Sampling and Identification
Between December 2022 and June 2023, a total of 150 clinical samples were collected, including wounds, burns, urine, ear swabs, and blood samples. Staphylococcus isolates were identified based on their growth on blood agar and mannitol salt agar. Characterization was carried out using conventional microbiological methods, including colonial morphology, Gram staining, and biochemical tests. Final identification was conducted using the VITEK 2 Compact system (bioMérieux, France), following the bacterial identification protocol cited previously [10].
2.2. Detection of Antibiotic Resistance Profile
The Kirby-Bauer disc diffusion method used 19 antibiotic discs (“Oxoid, UK”). The antibiotic susceptibility of the isolates was tested against a range of antibiotics, including amoxiclav, cefoxitin, methicillin, amoxicillin, ceftriaxone, piperacillin, ceftazidime, fusidic acid, tobramycin, tetracycline, gentamicin, azithromycin, vancomycin, clindamycin, lincomycin, ciprofloxacin, levofloxacin, trimethoprim-sulfamethoxazole, rifampicin, nitrofurantoin, meropenem, and chloramphenicol.
2.3. Biofilm Formation
Biofilm production was assessed using the microtiter plate method, as described by Yousefi et al. (2016).
2.4. Molecular Screening of mecA Gene
Bacterial DNA was extracted using the Promega DNA Mini Kit provided by Promega, USA. DNA was extracted from the bacteria, and its purity was measured. A specific gene, mecA, was then tested for forward “(5′-TCCAGATTACAACTTCACCAGG-3′)” and reverse “(5′-CCACTTCATATCTTGTAACG-3′)” primers, following the method described by Ghaznavi-Rad et al. The PCR mixture (20 µL total volume) included 10 µL master mix (Thermo Scientific, USA), 0.5 µL of each 10 µM primer, 1 µL DNA template, and 8 µL nuclease-free water. The PCR was carried out on a Bio-Rad MyCycler™ (USA) with the following settings: initial denaturation at 98°C for 30 seconds, 28 cycles of denaturation (98°C for 10 seconds), annealing (52°C for 30 seconds), and extension (72°C for 30 seconds), followed by a final extension at 72°C for (5 minutes) and a hold at 4°C. PCR products were analyzed by gel electrophoresis at 58 V for 120 minutes on a 1.4% agarose gel containing 0.5 µL gel stain (Bioteke, China). A 100 bp DNA ladder (Vivantis, Malaysia) served as a marker. The gel was visualized under UV light and imaged using a gel imager (Major Science, USA) [12].
2.5. Multilocus Sequence Typing
All strains were analyzed by the MLST protocol using primers listed in Tables 1 and 2. All PCR reactions were carried out in Applied Biosystems 2720 Thermal Cycler (USA).
| Gene Loci | Primer Sequence (5-3) | Amplicon Size |
|---|---|---|
| Arca | F AGTGACTCAAGTTGAA R AATCTTACCATCTAGG |
600 |
| SH 1200b | F CGGTAATGTAACACACGCAGT R TCTTCCTAGTAGCTGACCAG |
540 |
| HemHC | F CTGATCGTCAAGCTGAAGCAT R GTACCTGTGTGACCCTCAGA |
500 |
| leuBd | F AGCCATAGATTCGCATGGTGT R CCTAATGAACCTGGAATGGTAG |
650 |
| SH 1431e | F TCAGACCAATTCCCAACC R CTTTAGCGTCACGATGGTCG |
600 |
| CfxE f | F GAAGCACAAATTGATGGTCTGC R TCTGCCCCATTATCAACACA |
500 |
| Ribose ABC | F GAGACGATTCAGCTAAGCAA R CGCCTTTCATTAGGCCATTA |
650 |
| Gene | Primer sequence (5′–3′) | Amplicon (bp) |
| arcC |
F TGTGATGAGCACGCTACCGTTAG R TCCAAGTAAACCCATCGGTCTG |
508 |
| aroE |
F CATTGGATTACCTCTTTGTTCAGC “R CAAGCGAAATCTGTTGGGG” |
459 |
| gtr |
“F CAGCCAATTCTTTTATGACTTTT” “R GTGATTAAAGGTATTGATTTGAAT” |
508 |
| mutS |
F GATATAAGAATAAGGGTTGTGAA R GTAATCGTCTCAGTTATCATGTT |
608 |
| pyr |
F GTTACTAATACTTTTGCTGTGTTT R GTAGAATGTAAAGAGACTAAAATGAA |
851 |
| Tpi |
F ATCCAATTAGACGCTTTAGTAAC R TTAATGATGCGCCACCTACA |
592 |
| yqiL |
F CACGCATAGTATTAGCTGAAG R CTAATGCCTTCATCTTGAGAAATAA |
658 |
3. RESULTS AND DISCUSSION
3.1. Phenotypic Identification of Clinical Isolates
In the period from December 2022 to June 2023, 150 samples were collected from clinical sources, including patients of both sexes from different teaching hospitals and outpatients in Anbar/Iraq. All these samples were grown on mannitol salt agar and blood agar. The bacterial isolates included were 13.3% (n=20) S. epidermidis, 20% (n=30) S. haemolyticus, 23.3% (n=35) coagulase-positive staphylococci, 26% (n=39) Gram-negative bacteria, and 17.3% (n=26) showed no growth. Higher prevalence rates of methicillin-resistant coagulase-negative staphylococci (MRCoNS) have been reported in South Africa, reporting 86% in one study [14] and 100% in another study [15]. Among CoNS associated with infections, S. epidermidis and S. haemolyticus are recognized as significant etiological agents causing nosocomial infections [16]. Notably, S. epidermidis is the most frequently isolated staphylococcal species in humans and is considered the most clinically relevant CoNS species [17]. The highest percentage of coagulase-negative Staphylococcus (CoNS) was isolated from wound samples (36%), followed by burn samples (20%) and urine samples (18%). The lowest percentages were observed in ear swabs and blood samples, with 14% and 12%, respectively.
3.2. Antibiotic Sensitivity of Coagulase-Negative Staphylococcus
The antibiotic susceptibility testing of coagulase-negative Staphylococcus (CoNS) isolates showed the highest resistance rates to amoxiclav, cefoxitin, methicillin, amoxicillin, and ceftriaxone (100%). The isolates also exhibited high resistance to piperacillin (96%), ceftazidime (96%), fusidic acid (76%), tobramycin (68%), tetracycline (68%), gentamicin (66%), and azithromycin (66%). Resistance to vancomycin was observed in 52% of the isolates. Resistance levels were low against clindamycin (46%), lincomycin (42%), ciprofloxacin (42%), levofloxacin (40%), trimethoprim-sulfamethoxazole (38%), rifampicin (38%), and nitrofurantoin (18%). In comparison, the lowest resistance was observed against meropenem (10%) and chloramphenicol (2%). The results also confirmed that the study isolates were considered multidrug-resistant (MDR), with 76% showing resistance to six classes of antibiotics: penicillin, cephalosporins, macrolides, glycopeptides, tetracyclines, and fluoroquinolones.
An isolate is considered MDR when it shows resistance to at least three classes of antibiotic agents [18]. This virulent characteristic, i.e., its ability to resist multiple classes of antibiotics simultaneously, makes the bacterium one of the most intractable pathogens in the history of antibiotics [19]. MDR Staphylococcus can resist various agents through multiple mechanisms, such as altering the target site, producing deactivating enzymes, efflux pumping, and decreasing the intracellular concentration of antibiotics [20]. These mechanisms often arise due to prolonged or improper antibiotic use by patients [21]. This study revealed that meropenem is a highly effective antibiotic against multidrug-resistant coagulase-negative Staphylococcus. Meropenem acts on the cell wall like other beta-lactam antibiotics but differs in its high resistance to cephalosporinase or beta-lactamase enzymes [22].
However, chloramphenicol is the most effective antibiotic, exhibiting the highest efficacy. It is a broad-spectrum antibiotic that exerts its antibacterial effects by inhibiting bacterial protein synthesis at the ribosomal level. Its primary mechanism of action involves binding to the 50S ribosomal subunit, specifically at the peptidyl transferase center, to prevent the formation of peptide bonds between amino acids during translation, effectively halting polypeptide chain elongation. This inhibition disrupts bacterial protein synthesis, ultimately leading to bacteriostatic effects against most susceptible organisms [23]. The high susceptibilities recorded against these antibiotics could be due to the reserved use of those antibiotics, mainly for resistant staphylococcal infections. Thus, last-resort antibiotics still retain high activity against coagulase-negative Staphylococcus (CoNS) and may be used for empirical treatment of conditions, such as suspected CoNS sepsis, even though resistance to these antibiotics is gradually increasing [24]. The results showed that all isolates (100%) were methicillin-resistant coagulase-negative Staphylococcus. There was a complete concordance between phenotypic and genotypic results in confirming methicillin resistance in this study. This is primarily due to the presence of the mecA gene, which encodes the PBP2a protein that has a low affinity for beta-lactam antibiotics [25].
3.3. Biofilm Formation
The results of the microtiter plate (MTP) assay revealed that all the isolates demonstrated a positive ability for biofilm formation. Based on their biofilm-producing capacity, the isolates were classified into three categories: 58% (n=29) exhibited strong biofilm formation, 30% (n=15) displayed moderate biofilm production, and 12% (n=6) showed weak biofilm formation. These findings highlight the varying degrees of biofilm production among the isolates, which may have implications for their pathogenicity and persistence in clinical settings. Statistical analysis was performed to determine which clinical source was associated with the highest level of biofilm production and whether the observed differences were statistically significant. The results indicated that coagulase-negative Staphylococcus isolates from urine and wound sources exhibited significantly higher biofilm production compared to isolates from blood and ear swabs. Although biofilm production in burn isolates was higher than in those from blood and ear swab samples, the difference was not statistically significant (Table 3).
Isolates of Staphylococcus haemolyticus from urine and wound samples showed significantly higher biofilm production than isolates from blood and ear swabs. Burn isolates demonstrated elevated biofilm levels but without statistical significance. This suggests that specific clinical
| Comparison | P-value | Significance (p < 0.05) |
|---|---|---|
| Urine vs. Blood | < 0.01 | Significant |
| Wound vs. Blood | < 0.01 | Significant |
| Burn vs. Blood | 0.07 | Not Significant |
| Ear Swab vs. Blood | 0.08 | Not Significant |
| Urine vs. Wound | 0.90 | Not Significant |
| Urine vs. Burn | 0.04 | Significant |
environments may favor biofilm formation due to nutrient availability or local stress factors. Another study indicated that biofilm production was most prevalent in blood samples (27.3%), followed by pus swabs (12.5%) and pus aspirates (16.7%). Biofilm development is strongly associated with medical device-related infections, including contaminated implants, urinary catheters, and prosthetic valves [26]. Biofilm is an extracellular polysaccharide layer that facilitates bacterial attachment to surfaces and medical devices. S. haemolyticus isolates capable of forming biofilms contribute to infections associated with catheters and other medical devices [7]. S. epidermidis-producing biofilms can lead to bacteremia, especially in cases linked to catheter-associated and nosocomial infections. Biofilm formation by S. haemolyticus is a complex process that becomes more pronounced in the presence of antimicrobial agents [27].
3.4. Molecular Screening of mecA Gene
The results demonstrated that all the isolates were 100% positive for the detection of the mecA gene in methicillin-resistant coagulase-negative Staphylococcus isolates by using Uniplex PCR. Methicillin resistance occurs mainly because of a mutation in chromosome-encoded protein (penicillin-binding protein); bacteriophage transfers this mutation among CoNS isolates [28]. Methicillin-resistant strains have always exhibited a wide number of virulence factors and consequently show multi-drug resistance through different mechanisms [29]. In this study, the MRCoNS strains showed 100% multi-drug resistance, which is in agreement with the findings of a previous study [30].
The highest resistance rate of CoNS toward amoxiclav, cefoxitin, methicillin, amoxicillin, and ceftriaxone was observed. Methicillin resistance in isolates that lack the mecA gene may be mediated by other mechanisms of methicillin resistance, such as the presence of mecC and mecB genes [31]. The overproduction of β-lactamases, along with the emergence of methicillin resistance, has been documented in approximately 80% of CoNS species. This high prevalence significantly contributes to increased morbidity and mortality in hospital settings, as CoNS are major pathogens in healthcare-associated infections (HAIs) [24].
3.5. MLST Analysis
The molecular analysis identified five distinct sequence types (STs) of Staphylococcus epidermidis, demonstrating a high level of genotypic diversity. Notably, all isolates were recorded in Iraq for the first time, as presented in Table 4 and Fig. (1). ST89 was initially reported in Denmark in 1997 with ID 40259, with a global frequency of 27 isolates. In the current study, this ST has been identified in Iraq with ID 46590. Similarly, ST23 was first recorded in Mexico in 1996 with ID 40147 and has now been documented in Iraq with ID 46588. ST59 was originally detected in South Korea with ID 41234, and in this study, it has been identified in Iraq with ID 46587. This sequence type exhibited the highest frequency in this study, with a global occurrence of 101 isolates. Additionally, ST1183 was found to have the lowest global frequency, with only three recorded strains. These findings highlight the genetic diversity of S. epidermidis isolates and emphasize the importance of continued molecular surveillance to understand the epidemiology and distribution of this opportunistic pathogen. Moreover, there is a need for targeted public health measures and further research in Iraq. The Staphylococcus haemolyticus isolates analyzed in this study provide valuable insights into their global distribution and epidemiological significance. Sequence type (ST) 24 was first recorded in India in 2010 with ID 55, while in the current study, this isolate was identified in Iraq with ID 240. Similarly, ST35
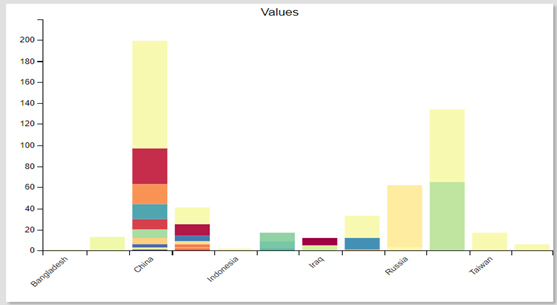
Shows the percentage distribution of Staphylococcus epidermidis isolates across Asia, highlighting regional variations. Notably, Iraq shows a significant proportion, possibly due to hospital-acquired infections, antimicrobial resistance, or diagnostic differences.
was initially reported in India in 2016 with ID 70 and has now been detected in Iraq with ID 241. ST109 was first identified in China in 2019 with ID 151, and in the present study, it has been recorded in Iraq with ID 242, with a frequency of two isolates. ST173 was initially reported in Burma in 2023 with ID 238 and has now been recorded in Iraq with ID 243. Likewise, ST146 was first documented in Thailand in 2021 with ID 197, and in the current study, this isolate has been detected in Iraq with ID 244. Other details of ST under study are listed in Table 4 and Fig. (2).
Alleles and sequence types (STs) were identified using the S. epidermidis-specific MLST database (https: //pubmlst. org/bigsdb?db =pubmlst_ sepidermidis _isolates& page=profiles &scheme _id=4) and for S. haemolyticus isolate, using its corresponding MLST database https: //pubmlst. org/bigsdb ?db=pubmlst _shaemolyticus _isolates &page= profiles &scheme _id=1).
| Isolates | ST | Methicillin Susceptibility | ID Profile | Isolation Area First Time | Source of Isolation | First Isolation Year in the World | Special ID for the Current Study | Frequency |
|---|---|---|---|---|---|---|---|---|
| 1. | 89 | *MRSE | 40259 | Denmark | blood | 1997 | 46590 | 27 isolates |
| 2. | 23 | MRSE | 40147 | Mexico | Blood | 1996 | 46588 | 60 isolates |
| 3. | 59 | MRSE | 41234 | South Korea | Wound swab | 2000 | 46587 | 101 isolates |
| 4. | 1183 | MRSE | 44665 | South Africa | CSF | 46591 | 3 isolates | |
| 5. | 35 | MRSE | 40165 | Portugal | Nasal swab | 2000 | 46592 | 22 isolates |
| 6. | 24 | *MRSH | 55 | India | Wound | 2010 | 240 | 2 isolates |
| 7. | 35 | MRSH | 70 | India | Wound | 2016 | 241 | 2 isolates |
| 8. | 109 | MRSH | 151 | China | UTI | 2019 | 242 | 2 isolates |
| 9. | 173 | MRSH | 238 | Burma | Wound | 2023 | 243 | 2 isolates |
| 10. | 146 | MRSH | 197 | Thailand | Wound | 2021 | 244 | 2 isolates |
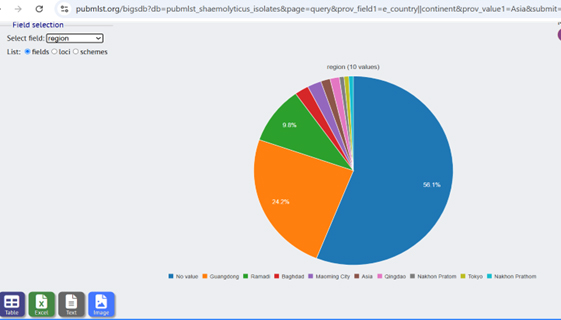
Shows the percentage distribution of Staphylococcus haemolyticus isolates across Asia, highlighting their prevalence in various countries. The green section represents the isolates recorded in Iraq, specifically in Ramadi, from the current study. This image presents a comparative analysis of the geographical spread of S. haemolyticus, emphasizing the newly identified isolates in Iraq and their contribution to the regional distribution.
The detection of various sequence types (STs) in Iraq suggests possible intercontinental transmission or convergent evolution, potentially driven by selective pressures in hospital environments. ST89 (from Denmark), ST23 (from Mexico), and the rare ST1183 indicate global dissemination and the importance of monitoring uncommon STs for antimicrobial resistance. For Staphylococcus haemolyticus, five distinct STs were identified, namely ST24 and ST35 (from India), ST109 (from China), ST173 (from Burma), and ST146 (from Thailand), indicating regional spread within Asia and broader international movement. This underscores the genetic diversity and global distribution of these pathogens. The emergence of these sequence types in Iraq may result from globalization, hospital transmission, and antimicrobial selective pressures [32]. The presence of globally distributed STs in S. epidermidis and S. haemolyticus suggests that healthcare-associated infections (HAIs) influence their population structure. As major opportunistic pathogens linked to bloodstream and prosthetic device infections with multidrug resistance, molecular surveillance is essential [33]. The detection of sequence types with varying global frequencies in Iraq highlights the need for continuous genetic monitoring to track bacterial evolution, antimicrobial resistance, and transmission pathways. Using MLST databases enabled precise classification and epidemiological assessment. Identifying both novel and known S. epidermidis and S. haemolyticus STs reveals their genetic diversity and clinical significance. Ongoing molecular surveillance is crucial to understand their distribution, resistance profiles, and pathogenic potential, supporting effective infection control strategies [34]. Another study indicated that MLST analysis of S. epidermidis identifies high-risk clones linked to nosocomial infections, tracks antibiotic-resistant strains, and guides infection control and antibiotic therapy. It enhances understanding of S. epidermidis epidemiology, supporting strategies to prevent and manage infections [35].
CONCLUSION
This study reveals high methicillin resistance, multidrug resistance (MDR), and strong biofilm formation in Staphylococcus epidermidis and Staphylococcus haemolyticus isolates from Anbar, Iraq. The identification of diverse sequence types (STs), reported for the first time in Iraq, highlights their genetic diversity and potential for intercontinental transmission. The presence of the mecA gene in all MRCoNS underscores the antimicrobial resistance threat. These findings emphasize the need for continuous molecular surveillance to track bacterial evolution and resistance, supporting effective infection control strategies and antibiotic stewardship in healthcare settings.
AUTHORS’ CONTRIBUTIONS
The authors confirm contribution to the paper as follows: H.K.B.: Study Concept or Design; A.H.A.: Data Curation; All authors reviewed the results and approved the final version of the manuscript.
LIST OF ABBREVIATIONS
| MLST | = Multilocus sequence typing |
| CoNS | = Coagulase-negative staphylococci |
| HAIs | = Healthcare-associated infections |
| MTP | = Microtiter plate |
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study obtained approval from the Ethics Committee of Al Anbar Medical Research University, Iraq, (approval number 23, December 5th, 2022).
HUMAN AND ANIMAL RIGHTS
All procedures performed in studies involving human participants were in accordance with the ethical standards of institutional and/or research committee and with the 1975 Declaration of Helsinki, as revised in 2013.
AVAILABILITY OF DATA AND MATERIALS
The authors confirm that the data supporting the findings of this research are available within the article.
ACKNOWLEDGEMENTS
Declared none.


